The gallery of photosynthetic reaction centers structures
Investigators: R. Pincak and M. Pudlak
Photosynthesis in bacteria
Photosynthetic Reaction Centers (RC's)
Light-Harvesting complex on bacterial photosynthesis
Photosynthesis in bacteria
Electron transfer through the RC's
References
PublicationsPhotosynthesis is a reaction in which light energy is converted into chemical energy. The primary process of photosynthesis is carried out by a pigment-protein complex embedded in the membrane, that is, reaction centers.
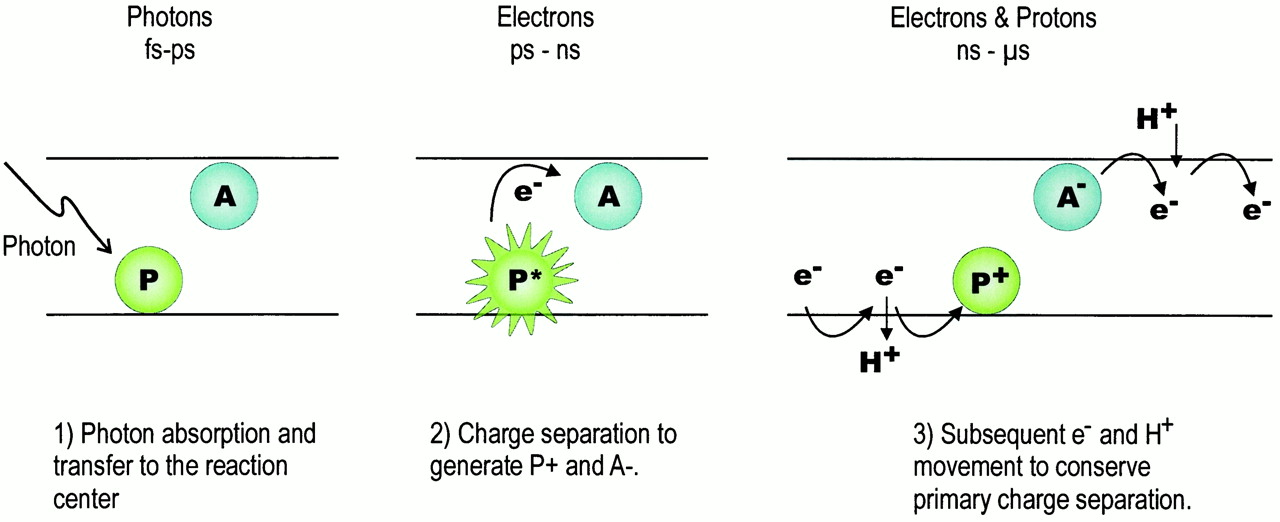
The photosynthetic reaction centers is a special pigment-protein complex, that functions as a photochemical trap. The function of the reaction center is to convert solar energy into biochemical amenable energy. Following the initial capture of a photon by antenna pigments, the photon is transferred to the RC pigments, where it gives rise to a separation and stabilization of charge across the photosynthetic membrane. Figure 1 depicts this process and illustrates the time scales typically involved.
Figure 1. Scheme of the primary processes in the photosynthetic RC. Here, P represents the charge-separating (bacterio) chlorophyll pigments (the primary electron donor) and A represents the first stable acceptor. Energy transfer from the antenna pigments leads to photoexcitation of P on the fs-ps time scale (left). Charge separation produces oxidized P+ and A- on the ps-ns scale (center). The recombination of P+A- to produce PA, heat and potentially damaging chemical species is efficiently prevented by further forward electron transfer that is now proton coupled. These more complex chemical processes ultimately produce stable photosynthetic products and occur, initially, on the ns and µs time scales (right)
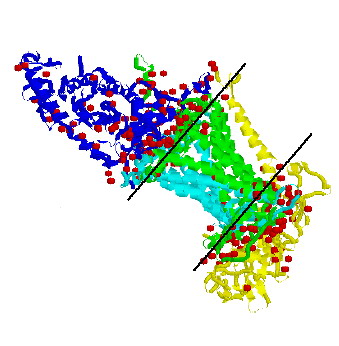
The photosynthetic reaction center (RC) is the first membrane protein whose three-dimensional structure was revealed at the atomic level by X-ray crystallograph more than fifteen years ago. Structural information about RC made a great contribution to the understanding of the reaction mechanism of the complicated membrane protein complex [5,6]. High-resolution structures of RC's from three photosynthetic bacteria are now available, namely, those from two mesophilic purple non-sulfur bacteria, Blastochloris viridis and Rhodobacter sphaeroides, and that from a thermophilic purple sulfur bacterium, Thermochromatium tepidum. In addition, a variety of structural studies, mainly by X-ray crystallography, are still being performed to give more detailed insight into the reaction mechanism of this membrane protein. The structural data from three RC's and their electron donors provided reliable models for molecular recognition in the primary step of bacterial photosynthesis.
Figure 2. Three-dimensional structure of tetrameric complex of RC penetrate through the membrane. Note that the water and Cytochrome molecules (blue, yellow color) do not penetrates the membrane spanning domain (black lines) of the reaction center complex. The chains of green and white blue color described M, L and H subunits of RC. (Cox&Lehninger, Principles of Biochemistry, Worth Publishing, 3rd ed, 2000).
Figure 3. X-ray structure arrangement of co-factors of RC embedded in the membrane protein complex (Cox &Lehninger, Principles of Biochemistry, Worth Publishing, 3rd ed, 2000).
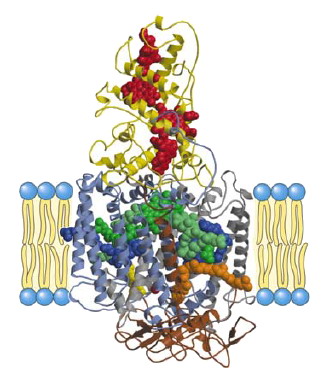
Electron transfer coupled with the uptake of protons across the membrane is a fundamental feature of bioenergetic processes such as oxidative phosphorylation and photosynthesis, and the resulting electrochemical gradient of protons is finally utilized for ATP synthesis (Fig. 8). Key players in bioenergetics are integral membrane proteins and co-factors embedded in the membrane protein complexes, where polypeptide chains spanning across the membrane provide a scaffold for the specific arrangement of co-factors in membrane protein complexes. Hence, structural data about membrane protein complexes contribute greatly to obtaining a profound understanding of reaction mechanisms [7].
In fact, a great deal of effort has been made to elucidate the tree-dimensional structures of membrane proteins involved in bioenergetics for the sake of functional analyses (Figs.2, 3). Of such structural studies, crystallographic studies of the photosynthetic reaction center (RC) provided the first successful description of the three-dimensional structure at an atomic resolution, and the methodology established in this structural work has had a strong influence on subsequent structural studies of membrane proteins.
In addition, structure analyses of RC complexes remain one of the most active fields in membrane protein structural biology. This is because it is not yet clear how the RC complex regulates the electrochemical properties of co-factors, especially that of the bacteriochlorophyll dimer (the special pair) that acts as the initiator of photosynthetic electron transfer, how the RC complex takes up protons to reduce quinone, which acts as the final electron acceptor in the complex, and how the RC complex accepts electrons from the electron carrier protein to reduce the photo-oxidized special-pair (Fig. 8). Consequently, structure analyses of some modified RC complexes, such as mutants or complexes with substrate analogues, have been carried out extensively so as to relate structural information to physical and chemical properties, in spite of the fact that the three-dimensional structure of the native RC complex has already been determined precisely.
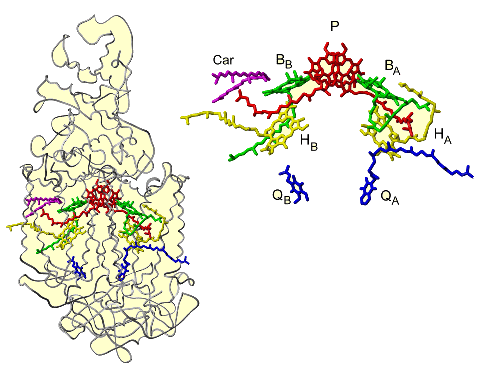
The RC complex maintains a number of prosthetic groups in the protein subunit scaffold. The prosthetic groups in the trans-membrane region apparently form two branches (A and B) that are related by a pseudo twofold axis perpendicular to the membrane plane.
These two branches run from the special-pair of bacteriochlorophyll (DA and DB) to the non-heme iron. Each branch consists of a bacteriochlorophyll monomer (BChlA or BChlB), bacteriopheophytin (BPhA or BPhB), and quinone (QA or QB). A carotenoid molecule is present in the trans-membrane region near BChlB. For more details on structural arrangement, see [10]. Branch A, mainly associated with the L subunit, is selectively utilized as the pathway of electron transfer, which is induced by charge-separation, in this process, an electron is emitted from the excited special-pair and transferred through BPhA and QA [11,12]. The involvement of BChlA, which is located between the special-pair and BPhA, in the electron transfer remains a matter of debate. Branch B, is associated with the M subunit and is inactive in electron transfer. QB is the final electron acceptor at this stage, and the reduced QB molecule serves as the electron carrier to the Cyt bc1 complex. The RC structures from photosynthetic bacteria are described schematically in Figs.2-4.
Figure 4. Schematic representation of the X-ray structure of Rp. viridis RC showing two symmetrical subunits of the RC
Since prosthetic groups play a central role in photosynthetic energy conversion, it is necessary to describe the three-dimensional arrangement of these prosthetic groups and their interactions with protein subunits precisely, this will lead to a better understanding of the functional aspects of RC and probably give answer to the questions why electron transfer through the RC prefers only one branch(L)
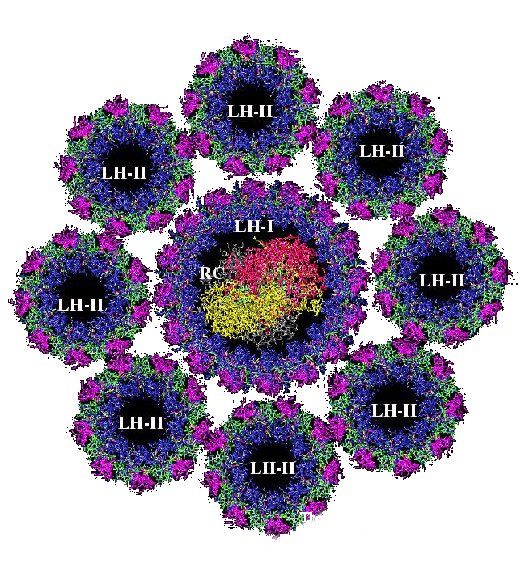
Photochemistry begins at the (bacterial) reaction center where charge is separated across the membrane. The reaction center (RC) molecule requires energy to perform this task, either from direct absorption of a photon, or energy transferred from a light-harvesting complex. All purple bacteria produce a primary light-harvesting complex (LH1), which is intimately associated with the RC - this composite is termed the Core complex. Most purple bacteria produce a peripheral light-harvesting complex (LH2), and some produce an additional peripheral complex (sometimes called LH3)(Fig. 5).
Figure 5. Crystal structure of an integral membrane light-harvesting complex from photosynthetic bacteria (http://www.chem.gla.ac.uk/protein/LH2/core.html).Each light-harvesting complex is an oligomer formed from a subunit consisting of two or more polypeptide chains with associated pigment molecules [1]. The pigments used in LH complexes are bacteriochlorophyll a and carotenoid. BChlA is the primary pigment, carotenoid molecules are primarily used for photoprotection, although they do function as additional light-harvesting pigments.
Primary absorption of a photon light is absorbed either by bacteriochlorophylls or carotenoids in different spectral regions. Two kinds of bacteriochlorophylls absorb at slightly different energies and at different angles. The ring structure enhances absorption and generates an energy trap. Excitation Transfer between Light-Harvesting Complexes. The coplanar arrangement of BChls is ideal for excitation transfer between the BChl rings which proceeds within few picoseconds. Transfer from the LH-I ring to the RC is accelerated by bridge BChls. Bacterial RCs accept light energy from light-harvesting antenna proteins, referred to as LHI and LHII. In the photosynthetic apparatus, the RC complex is surrounded by the LHI-ring.
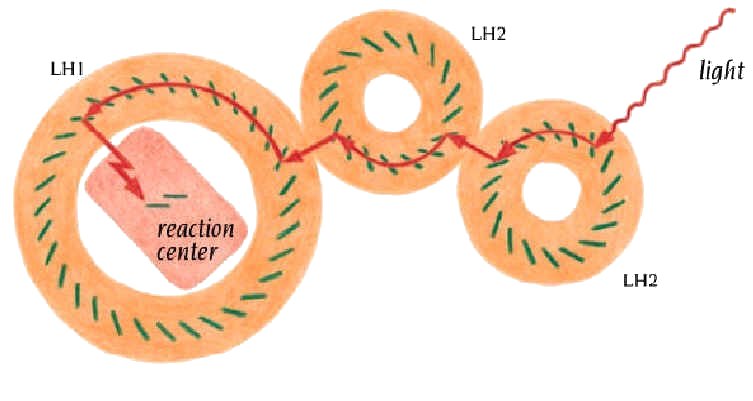
The protein matrix which supports these molecules in each of the distinct LH complexes modulates the absorption of the chromophores. This modulation results in a downward gradient of energy levels from LH3 - LH2 - LH1 (Fig.6). The bacteria synthesize enough LH2 to satisfy the RC, this is directly dependent on the ambient light intensity, the number of BChlA per RC rises to 250 or more in low light cases.
Figure 6. Light harvesting complex and light capturing pathway from LH2 to reaction center (Branden&Tooze, Introduction to Protein Structure, 2nd ed, Garland publishing, 1999).
Purple bacteria have been extensively studied as a means to understanding the processes involved in photosynthesis. Their light-harvesting systems possess, relatively, many residues per pigment, and the spectra of these bacterial systems are relatively straightforward: they posses only one major chlorin type pigment and the resonant absorption bands arising from this chromophore are generally well resolved. This makes this bacterial system ideal as a model for studying photosynthesis in general.
It is through photosynthesis that earth's biosphere derives its energy from sunlight. Photosynthetic organisms, i.e., plants, algae and photosynthetic bacteria, have developed efficient systems to harvest the light of the sun and to use the light energy to drive their metabolic reactions, such as the reduction of carbon dioxide to sugar. The ubiquitous green color of plants is testimony to the key molecular participant in the light harvesting of plants, chlorophylls. More hidden in this respect, but no less widespread, is a second participating molecule, carotenoid. In green leaves the color of the carotenoids is masked by the much more abundant chlorophylls while in red ripe tomatoes or petals of yellow flowers, the carotenoids predominate. Chlorophyll molecules exist in slightly different chemical structures in various photosynthetic organisms, as chlorophyll a or b in plants or algae, and as bacteriochlorophyll a (BChlA) or b in photosynthetic bacteria. Molecules such as chlorophyll and carotenoid that absorb light and impart color to living matter and other materials are called pigments.
In general, biological pigments are non-covalently bound to proteins, forming the so-called pigment-protein complexes. The pigment-protein complexes are organized as the photosynthetic unit (PSU). The bacterial PSU consists of two types of pigment-protein complexes: the photosynthetic reaction centers (RCs) and the light-harvesting complexes. The main function of the light-harvesting complexes is to gather light energy and to transfer this energy to the reaction centers for the photo-induced redox processes (Fig.7).
Figure 7. Antenna or light harvesting complex distribution of RC's and LH complex promoting multiple photon absorption (Voet&Voet, Biochemistry, 2nd ed, John Wiley&Sons, 1995).
Purple bacteria are great masters of harvesting light. Nearly all the energy gained by the absorption of a photon is transferred on to the reaction center. The purple bacteria exploit elegant quantum physics, the working of which were only fully understood recently after the discovery of the structures of light-harvesting complexes and investigations into their electronic excitations [3].
In photosynthetic purple bacteria, the cyclic electron transfer reaction is performed by RC and two other components: the cytochrome (Cyt) bc1 complex, and the soluble
electron carrier protein (Fig. 8).
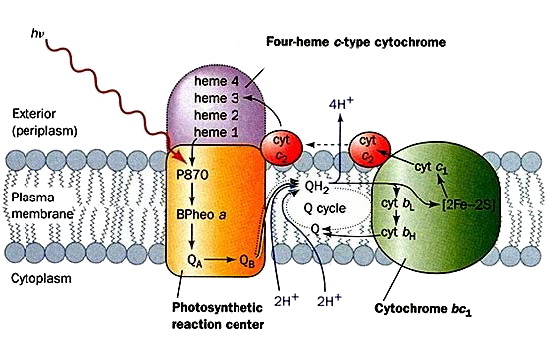
Figure 8 describes in purple bacteria that the primary reactions of photosynthesis are performed on the inner membrane, referred to as the photosynthetic membrane. First, RC accepts light energy from light harvesting antenna complexes, LH-I and LH-II. The RC complex contains various prosthetic groups that serve as the photosynthetic pigments, such as the bacteriochlrophyll dimer (special-pair), bacteriopheophytin, and quinone; charge separation occurs in this RC complex. As a result of the charge separation, quinone is reduced to quinol. Second, quinol moves to the Cyt bc1 complex through the membrane. The Cyt bc1 complex re-oxidizes quinol to quinone, and transfers electrons to the soluble electron carrier proteins. The soluble electron carrier proteins are classified into two groups, Cyt c2 and the high-potential iron-sulfur protein (HiPIP), depending on the species used physiologically. In each case, the soluble electron carrier protein contains a redox center, such as a heme c group or an Fe-S cluster. Finally, the soluble electron carrier proteins move through the periplasmic space, and transfer electrons to RC. The photo-oxidized special-pair is reduced, and RC returns to the initial state. In the course of this cyclic electron transfer, the oxidation and reduction of quinone bring about a trans-membrane electrochemical gradient of protons, and the resulting energy is utilized for ATP synthesis by ATP synthase. The electron flow, proton transfer, the absorption of light energy and ATP synthesis are represented by arrows (Messerschmidt&Huber, Handbook of Metalloproteins}, John Wiley&Sons, Ltd, Chichester, 2001).
A critical aspect of the photochemistry of reaction centers is their ability to perform electron transfer (ET) with a quantum yield of almost unity. Within 30 ps after excitation a stable charge separated state is formed in all photosystems. Although the nature of the spectral changes with time is complex, the role of many factors driving the initial electron transfer process has been established in purple bacteria. For other types of reaction centers, the larger number of tetrapyrroles and the highly overlapping nature of the optical bands have hindered interpretation of the optical changes, and research continues to delineate the electron transfer processes.
After extensive studies, the electron rate in purple bacteria is now established to be critically coupled to the properties of the bacteriochlorophyll monomer that lies between the donor and bacteriopheophytin acceptor (Fig. 4). The involvement of the bacteriochlorophyll monomer may give rise to multiple pathways for electron transfer [13] and can partially determine the asymmetry of the electron transfer along one branch [14].
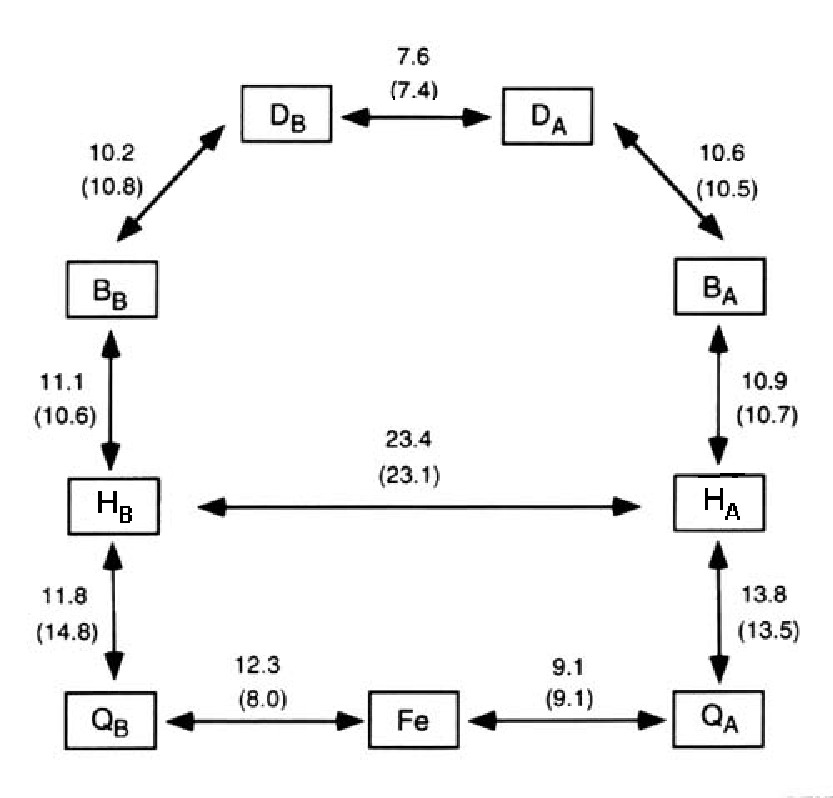
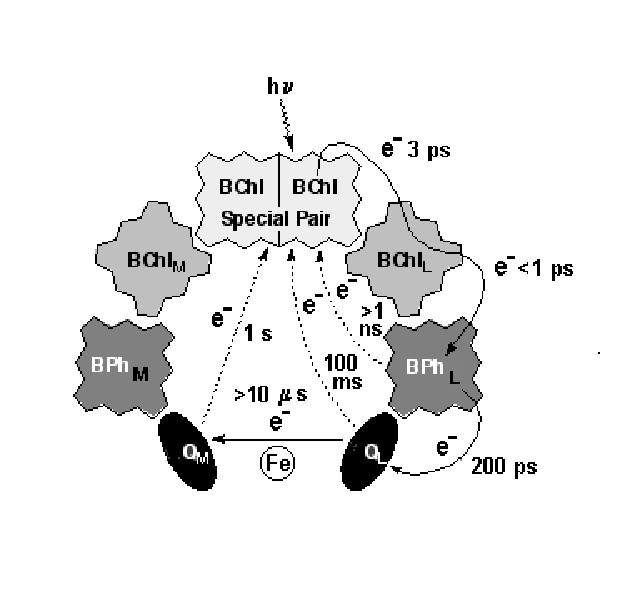
Despite the striking symmetry of the cofactors into 2 branches (Fig.4), electron transfer in the pheophytin-quinone reaction centers proceeds only along one branch (L) with at least a 10:1 ratio. Typical distance and electron transfer rate between cofactors in the RC of Purple Bacteria are shown in Figs.9,10.
Figure 9. Distances (in A) between the cofactors in the RC's of Rb. sphaeroides and of Rp. viridis (in brackets) using only the porphyrin and the quinone rings for the centre of mass calculations.
Figure 10. Charge-separated intermediates and their lifetimes in the bacterial photosynthetic reaction center.
In plants and bacteria the energy of light is stored in the energy of the electric potential later used to form chemical bonds. The reaction center complex from the anoxygenic purple photosynthetic
bacteria are the best understood of all photosynthetic organisms, from both a structural and a functional point of view. Photosynthesis begins when light is absorbed by an antenna pigment. Antennas permit an organism to increase greatly the absorption cross section for light without having to build an entire reaction center and associated electron transfer system for each pigment, which would be very costly in terms of cellular resources. The excited (bacterio)chlorophyll molecule transfers an electron to a nearby acceptor molecule, thereby creating an ion pair state consisting of the oxidized chlorophyll and reduced acceptor. After the initial electron transfer event, a series of electron transfer reactions takes place that eventually stabilizes the stored energy in forms that can be used by the cell. Open areas of investigation are delineation of the mechanism of electron transfer from the primary to the secondary quinone and the role of quinone movement during electron transfer [15,16].Our understanding of the primary processes in photosynthesis is not complete without explanation of the strong asymmetry in ET. We believe that the reason for asymmetric ET between prosthetic groups located on different polypeptides is a different molecular dynamics. Dynamics of atoms causes the change of the electrical potential fields and the conformational variations influence the mutual orientations between cofactors. Then the energy gap and overlap of electronic wave functions fluctuates as a result in the system. The net result is a different fluctuation of electronic energy levels on prosthetic groups and also a different fluctuations of the overlaps of the electronic wave functions on L and M branches. On the other hand the chain located on M is inactive in ET and the highly asymmetric functionality, however, can be decreased by amino acid mutations or cofactor modification. This approach can be used to explain the effect of individual amino acid mutation or cofactor modifications on the observed balance between the forward ET reaction on the L-side of the RC, the charge recombination processes, and ET to the M-side chromophores [17,18,19,20].